If you want to use acoustic parameters extracted by the wonderful Praat software with nkululeko, you state
[FEATS]
type=['praat']
in the feature section of your config file.
If you like to use only some features of all the ones that are extracted by David R. Feinberg's Praat scripts, you can look at the output and select some of them in the FEAT section, e.g.
type = ['praat']
praat.features = ['speechrate(nsyll / dur)']
You can do the same with opensmile features:
type = ['os']
os.features = ['F0semitoneFrom27.5Hz_sma3nz_amean']
or even combine them
type = ['praat', 'os']
praat.features = ['speechrate(nsyll / dur)']
os.features = ['F0semitoneFrom27.5Hz_sma3nz_amean']
this is actually the same as
type = ['praat', 'os']
features = ['speechrate(nsyll / dur)', 'F0semitoneFrom27.5Hz_sma3nz_amean']
if you would want to combine all of opensmile eGeMAPS features with selected Praat features, you would do:
type = ['praat', 'os']
praat.features = ['speechrate(nsyll / dur)']
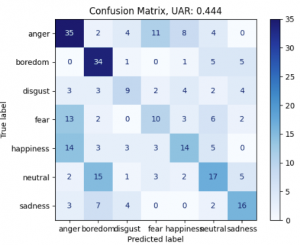
It is interesting to see, how many emotions of Berlin EmoDB still get recognized with only mean F0 and Jitter as features:

What kind of features are there, you might ask yoursel?
Here's a list:
'duration', 'meanF0Hz', 'stdevF0Hz', 'HNR', 'localJitter',
'localabsoluteJitter', 'rapJitter', 'ppq5Jitter', 'ddpJitter',
'localShimmer', 'localdbShimmer', 'apq3Shimmer', 'apq5Shimmer',
'apq11Shimmer', 'ddaShimmer', 'f1_mean', 'f2_mean', 'f3_mean',
'f4_mean', 'f1_median', 'f2_median', 'f3_median', 'f4_median',
'JitterPCA', 'ShimmerPCA', 'pF', 'fdisp', 'avgFormant', 'mff',
'fitch_vtl', 'delta_f', 'vtl_delta_f''