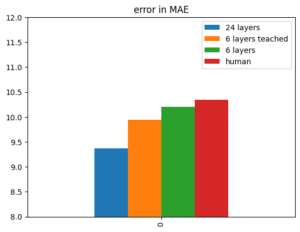
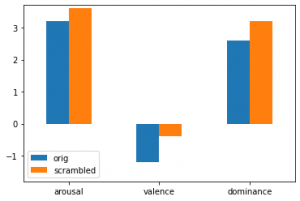
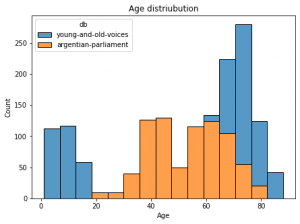
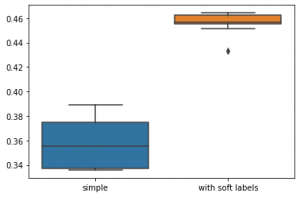
This post shows you how to classify emotions with a Multi Layer Perceptron (MLP) artificial neural net based on the torch framework (a different very famous ANN framework would be Keras).
Here's a complete jupyter notebook for your convenience.
We start with some imports, you need to install these packages, e.g. with pip, before you run this code:
import audformat
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import torch
import os
import opensmile
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import recall_score
Then we need to download and prepare our sample dataset, the Berlin emodb:
# get and unpack the Berlin Emodb emotional database if not already there
if not os.path.isdir('./emodb/'):
!wget -c https://tubcloud.tu-berlin.de/s/8Td8kf8NXpD9aKM/download
!mv download emodb_audformat.zip
!unzip emodb_audformat.zip
!rm emodb_audformat.zip
# prepare the dataframe
db = audformat.Database.load('./emodb')
root = './emodb/'
db.map_files(lambda x: os.path.join(root, x))
df_emotion = db.tables['emotion'].df
df = db.tables['files'].df
# copy the emotion label from the the emotion dataframe to the files dataframe
df['emotion'] = df_emotion['emotion']
As neural nets can only deal with numbers, we need to encode the target emotion labels with numbers:
# Encode the emotion words as numbers and use this as target
target = 'enc_emo'
encoder = LabelEncoder()
encoder.fit(df['emotion'])
df[target] = encoder.transform(df['emotion'])
Now the dataframe should look like this:
df.head()

To ensure that we learn about emotions and not speaker idiosyncrasies we need to have speaker disjunct training and development sets:
# define fixed speaker disjunct train and test sets
train_spkrs = df.speaker.unique()[5:]
test_spkrs = df.speaker.unique()[:5]
df_train = df[df.speaker.isin(train_spkrs)]
df_test = df[df.speaker.isin(test_spkrs)]
print(f'#train samples: {df_train.shape[0]}, #test samples: {df_test.shape[0]}')
#train samples: 292, #test samples: 243
Next, we need to extract some acoustic features:
# extract (or get) GeMAPS features
if os.path.isfile('feats_train.pkl'):
feats_train = pd.read_pickle('feats_train.pkl')
feats_test = pd.read_pickle('feats_test.pkl')
else:
smile = opensmile.Smile(
feature_set=opensmile.FeatureSet.GeMAPSv01b,
feature_level=opensmile.FeatureLevel.Functionals,
)
feats_train = smile.process_files(df_train.index)
feats_test = smile.process_files(df_test.index)
feats_train.to_pickle('feats_train.pkl')
feats_test.to_pickle('feats_test.pkl')
Because neural nets are sensitive to large numbers, we need to scale all features with a mean of 0 and stddev of 1:
# Perform a standard scaling / z-transformation on the features (mean=0, std=1)
scaler = StandardScaler()
scaler.fit(feats_train)
feats_train_norm = pd.DataFrame(scaler.transform(feats_train))
feats_test_norm = pd.DataFrame(scaler.transform(feats_test))

Next we define two torch dataloaders, one for the training and one for the dev set:
def get_loader(df_x, df_y):
data=[]
for i in range(len(df_x)):
data.append([df_x.values[i], df_y[target][i]])
return torch.utils.data.DataLoader(data, shuffle=True, batch_size=8)
trainloader = get_loader(feats_train_norm, df_train)
testloader = get_loader(feats_test_norm, df_test)
We can then define the model, in this example with one hidden layer of 16 neurons:
class MLP(torch.nn.Module):
def __init__(self):
super().__init__()
self.linear = torch.nn.Sequential(
torch.nn.Linear(feats_train_norm.shape[1], 16),
torch.nn.ReLU(),
torch.nn.Linear(16, len(encoder.classes_))
)
def forward(self, x):
# x: (batch_size, channels, samples)
x = x.squeeze(dim=1)
return self.linear(x)
We define two functions to train and evaluate the model:
def train_epoch(model, loader, device, optimizer, criterion):
model.train()
losses = []
for features, labels in loader:
logits = model(features.to(device))
loss = criterion(logits, labels.to(device))
losses.append(loss.item())
optimizer.zero_grad()
loss.backward()
optimizer.step()
return (np.asarray(losses)).mean()
def evaluate_model(model, loader, device, encoder):
logits = torch.zeros(len(loader.dataset), len(encoder.classes_))
targets = torch.zeros(len(loader.dataset))
model.eval()
with torch.no_grad():
for index, (features, labels) in enumerate(loader):
start_index = index * loader.batch_size
end_index = (index + 1) * loader.batch_size
if end_index > len(loader.dataset):
end_index = len(loader.dataset)
logits[start_index:end_index, :] = model(features.to(device))
targets[start_index:end_index] = labels
predictions = logits.argmax(dim=1)
uar = recall_score(targets.numpy(), predictions.numpy(), average='macro')
return uar, targets, predictions
Next we initialize the model and set the loss function (criterion) and optimizer:
device = 'cpu'
model = MLP().to(device)
criterion = torch.nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=0.0001)
epoch_num = 250
uars_train = []
uars_dev = []
losses = []
We can then do the training loop over the epochs:
for epoch in range(0, epoch_num):
loss = train_epoch(model, trainloader, device, optimizer, criterion)
losses.append(loss)
acc_train = evaluate_model(model, trainloader, device, encoder)[0]
uars_train.append(acc_train)
acc_dev, truths, preds = evaluate_model(model, testloader, device, encoder)
uars_dev.append(acc_dev)
# scale the losses so they fit on the picture
losses = np.asarray(losses)/2
Next we might want to take a look at how the net performed with respect to unweighted average recall (UAR):
plt.figure(dpi=200)
plt.plot(uars_train, 'green', label='train set')
plt.plot(uars_dev, 'red', label='dev set')
plt.plot(losses, 'grey', label='losses/2')
plt.xlabel('eopchs')
plt.ylabel('UAR')
plt.legend()
plt.show()

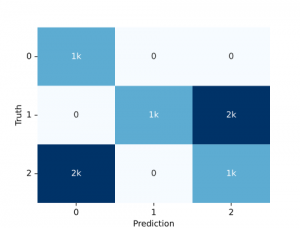
And perhaps see the resulting confusion matrix:
from sklearn.metrics import ConfusionMatrixDisplay
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(truths, preds, normalize = 'true')
disp = ConfusionMatrixDisplay(confusion_matrix=cm, display_labels=encoder.classes_).plot(cmap='gray')