audEERING recently published an emotion prediction model based on a finetuned Wav2vec2 transformer model.
Here I'd like to show you how you can use this model to predict your audio samples (it is actually also explained in the Github link above).
As usual, you should start with dedicating a folder on your harddisk for this and install a virtual environment:
virtualenv -p=3 venv
which means we want python version 3 (and not 2)
Don't forget to activate it!
Then you would need to install the packages that are used:
pandas
numpy
audeer
protobuf == 3.20
audonnx
jupyter
audiofile
audinterface
easiest to copy this list into a file called requierments.txt and then do
pip install -r requirements.txt
and start writing a python script that includes the packages:
import audeer
import audonnx
import numpy as np
import audiofile
import audinterface
, load the model:
# and download and load the model
url = 'https://zenodo.org/record/6221127/files/w2v2-L-robust-12.6bc4a7fd-1.1.0.zip'
cache_root = audeer.mkdir('cache')
model_root = audeer.mkdir('model')
archive_path = audeer.download_url(url, cache_root, verbose=True)
audeer.extract_archive(archive_path, model_root)
model = audonnx.load(model_root)
sampling_rate = 16000
signal = np.random.normal(size=sampling_rate).astype(np.float32)
load a test sentence (in 16kHz 16 bit wav format)
# read in a wave file for testing
signal, sampling_rate = audiofile.read('test.wav')
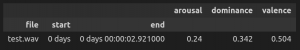
and print out the results
# print the results in the order arousal, dominance, valence.
print(model(signal, sampling_rate)['logits'].flatten())
You can also use audinterace's magic and process a whole list of files like this:
# define the interface
interface = audinterface.Feature(
model.labels('logits'),
process_func=model,
process_func_args={
'outputs': 'logits',
},
sampling_rate=sampling_rate,
resample=True,
verbose=True,
)
# create a list of audio files
files = ['test.wav']
# and process it
interface.process_files(files).round(2)
should result in:
Also check out this great jupyter notebook from audEERING