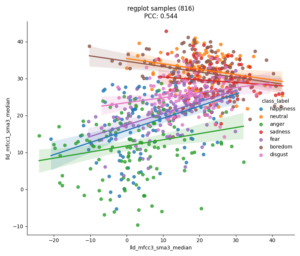
With nkululeko since version 0.77.7 there is a new interface named multidb which lets you compare several databases.
You can state their names in the [EXP] section and they will then be processed one after each other and against each other, the results are stored in a file called heatmap.png in the experiment folder.
!Mind YOU NEED TO OMIT THE PROJECT NAME!
Here is an example for such an ini.file:
[EXP]
root = ./experiments/emodbs/
# DON'T give it a name,
# this will be the combination
# of the two databases:
# traindb_vs_testdb
epochs = 1
databases = ['emodb', 'polish']
[DATA]
root_folders = ./experiments/emodbs/data_roots.ini
target = emotion
labels = ['neutral', 'happy', 'sad', 'angry']
[FEATS]
type = ['os']
[MODEL]
type = xgb
you can (but don't have to), state the specific dataset values in an external file like above.
data_roots.ini:
[DATA]
emodb = ./data/emodb/emodb
emodb.split_strategy = specified
emodb.test_tables = ['emotion.categories.test.gold_standard']
emodb.train_tables = ['emotion.categories.train.gold_standard']
emodb.mapping = {'anger':'angry', 'happiness':'happy', 'sadness':'sad', 'neutral':'neutral'}
polish = ./data/polish_emo
polish.mapping = {'anger':'angry', 'joy':'happy', 'sadness':'sad', 'neutral':'neutral'}
polish.split_strategy = speaker_split
polish.test_size = 30
Withe respect to the mapping, you can also specify super categories, by giving a list as a source category. Here's an example:
emodb.mapping = {'anger, sadness':'negative', 'happiness': 'positive'}
labels = ['negative', 'positive']
Call it with:
python -m nkululeko.multidb --config my_conf.ini
The default behavior is that all databases are used as a whole when being test or training database. If you would rather like the splits to be used, you can add a flag for this:
[EXP]
use_splits = True
Here's a result with two databases:

and this is the same experiment, but with augmentations:

In order to add augmentation, simply add an [AUGMENT] section:
[EXP]
root = ./experiments/emodbs/augmented/
epochs = 1
databases = ['emodb', 'polish']
[DATA]
--
[AUGMENT]
augment = ['traditional', 'random_splice']
[FEATS]
...
In order to add an additional training database to all experiments, you can use:
[CROSSDB]
train_extra = [meta, emodb]
, to add two databases to all training data sets,
where meta and emodb should then be declared in the root_folders file