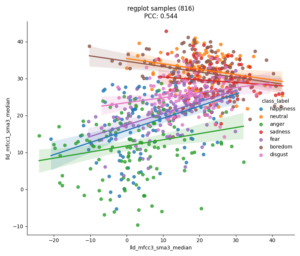
With nkululeko since version 1.0.1 we integrated a text classification model. It's a so-called zero-shot model, which means you can define the categories you would like to have predicted by yourself.
Prerequisite for this is that your data is transcribed, i.e. there is a text column in your data.
Here is an example ini file how to use this on a transcripted version of emodb
[EXP]
root = ./examples/results
name = emodb_textclassifier
[DATA]
databases = ['emodb']
emodb = ./examples/results//exp_emodb_translate/results/all_predicted.csv
emodb.type = csv
emodb.split_strategy = random
labels = ['anger', 'happiness']
target = emotion
[FEATS]
type = ['os']
store_format = csv
[MODEL]
type = svm
[PREDICT]
targets = ['textclassification']
textclassifier.candidates = ["sadness", "anger", "neutral", "happiness", "fear", "disgust", "boredom"]
The output is a version with all columns and one with only the pewdicted emotions (from text)
file,start,end,classification_winner,sadness,anger,neutral,happiness,fear,disgust,boredom
./data/emodb/emodb/wav/12a01Fb.wav,0 days,0 days 00:00:01.863625,neutral,0.11576763540506363,0.1414959877729416,0.3593694567680359,0.05933323875069618,0.08951663225889206,0.12100014835596085,0.11351688951253891
./data/emodb/emodb/wav/12a01Wc.wav,0 days,0 days 00:00:02.358812500,neutral,0.12048673629760742,0.1446247100830078,0.25808465480804443,0.04279503598809242,0.0794658437371254,0.25803136825561523,0.09651164710521698
It makes sense that almost all predicted labels are neutral, because emodb was designed to have linguistically neutral emotional content.
Following the winner class are the logits for all candidate classes.