This is a tutorial on how to
- configure a python environment with Jupyter notebook
- download Berlin EmoDB
- import the audformat database
- extract acoustic features with opensmile
- perform a machine classification with sklearn
It does expect some experience with
- unix commands
- python
- pandas
So if you miss this you might have to google the stuff you don't understand.
In case you know German and use Windows I recorded this screencast for you.
There is a Kaggle notebook that you could use to try this out.
Configure a python environment
I start from the point where you got python installed on your machine and have a shell (console window).
I use Unix commands here, most of them should also work on Mac OS, for Windows you might have to adapt some (e.g. 'ls' becomes 'dir').
So if you type
python3
in your shell, the python interpreter should start, you can quit it with the command
exit()
Create a subfolder for your project and enter it, e.g.
mkdir emodb; cd emodb
Create a virtual environment for your project
python3 -m venv ./venv
Activate your project
source ./venv/bin/activate
which should result in your prompt including the environment name like e.g. this
(venv)>
You can leave your environment with the
deactivate
command. For now though, please make sure you have the environment activated. You can then install the most important packages with pip like this:
pip install pandas numpy jupyter audformat opensmile sklearn matplotlib
If all goes well, you should now be able to start up the jupyter server which should give you an interface in your browser window.
jupyter notebook &
And create a new notebook by clicking the "New" button near the left top corner.
Get and unpack the Berlin Emodb emotional database
You would start by downloading and unpacking emodb like this (of course you can do this as well outside the notebook in your shell):
!wget -c https://tubcloud.tu-berlin.de/s/8Td8kf8NXpD9aKM/download
!mv download emodb_audformat.zip
!unzip emodb_audformat.zip
!rm emodb_audformat.zip
with
import audformat
db = audformat.Database.load('./emodb')
db
you can load the database and inspect the contents.
You still have to state the absolute path to the audio files for further processing. You would find the current directory with the
!pwd
command, and would add the emodb folder name to it and prefix this to the wav file paths like so
import os
root = '/...my current directory.../emodb/'
db.map_files(lambda x: os.path.join(root, x))
To check that this worked you might want to listen to a sample file
import IPython
IPython.display.Audio(db.tables['emotion'].df.index[0])
which should give you a GUi like this

Extract acoustic features
EmoDB is annotated with emotional labels. If we want to classify these emotions automatically we need to extract acoustic features first.
We can do this easily in python with dedicated packages for this like the Praat software or opensmile. In this tutorial we'll use opensmile.
First we will get the Pandas dataframes from the database like this:
df_emo = db.tables['emotion'].df
df_files = db.tables['files'].df
and might want to inspect the class distribution with pandas
df.emotion.value_counts().plot(kind='pie')
then, with
import opensmile
smile = opensmile.Smile(
feature_set=opensmile.FeatureSet.GeMAPSv01b,
feature_level=opensmile.FeatureLevel.Functionals,
)
you construct your feature extractor and with
df_feats = smile.process_files(df_emo.index)
should be able to extract the 62 GeMAPS acoustic features, which you could check by looking at the dimension of the dataframe
df_feats.shape
and looking at the first entry
df_feats.head(n=1)
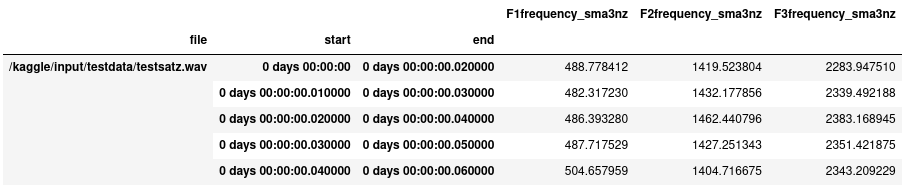
You might run into trouble later because the smile.process function per default results in a multiindex with filename, start and end time (because you might have extracted low level features per frame). In the following picture i show my screen so far to illustrate the situation.

So you end up with three data frames:
Perform a statistical classification on the data
Now we would conclude this tutorial by performing a first machine classification.
You basically need four sets of data for this: each a feature and label set for a training and a test (or better: development) set.
In a naive approach, we use the first 100 entries of the EmoDB for test and the others for training:
test_labels = df_emo.iloc[:100,].emotion
train_labels = df_emo.iloc[100:,].emotion
test_feats = df_feats.iloc[:100,]
train_feats = df_feats.iloc[100:,]
There are numerous possibilities to use machine classifiers in python, if we don't want to code one ourselves we might want to use on from the sklearn package, for example an implementation of the SVM (support vector machine) algorithm
from sklearn import svm
clf = svm.SVC()
train it with our training features and labels
clf.fit(train_feats, train_labels)
, compute predictions on the test features
pred_labels = clf.predict(test_feats)
and compare the predictions with the real labels (aka ''ground truth'') with a confusion matrix
from sklearn.metrics import confusion_matrix
confusion_matrix(test_labels.emotion, pred_labels)
and by computing the unweighted average recall (UAR)
from sklearn.metrics import recall_score
recall_score(test_labels, pred_labels, average='macro')
This results in chance level as the SVM classifier lazily always decided on the majority class. The results can be improved to something more meaningful, e.g. by passing better meta parameters when constructing the classifier:
clf = svm.SVC(kernel='linear', C=.001)
and repeating the experiment, which should result in a confusion matrix like this one (see Kaggle notebook for code):
 .
.
This concludes the tutorial so far, what to do next?
Here are some suggestions:
- What is really problematic with the above approach is that the training and the test set are not speaker independent, i.e. the same 10 speakers appear in both sets.
- Which means you can not know if the classifier learned anything about emotions or (more probable) some idiosyncratic peculiarities of the speakers.
- With so few speakers it doesn't make a lot of sense to further divide them, so what people often do is perform a LOSO (leave-one-speaker-out) or do x-cross validation by testing x times a part of the speakers against the others (in the case of EmoDB this would be the same if x=10).
- What's also problematic is that you only looked at one (very small, highly artificial) database and this usually does not result in a usable model for human emotional behavior.
- Try to import a different database or record your own, map the emotions to the EmoDB set and see how this performs.
- SVMs are great, but you might want to try other classifiers.
- Perform a grid search on the best meta-parameters for the SVM.
- Try other sklearn classifiers.
- Try other famous classifiers like e.g. XGBoost.
- Try ANNs (artificial neural nets) with keras or torch.
- Try other features
- There are other opensmile feature set configurations available.
- Do feature selection and to identify the best ones to see if they make sense.
- Try other features, e.g. from Praat or other packages.
- Try embeddings from pretrained ANNs like e.g. Trill or PANN features.
- The opensmile features are all given as absolute values.
- Try to normalize them with respect to the training set or each speaker individually.
- Generalization is often improved by adding acoustic conditions to the training:
- Try augmenting the data by adding samples mixed with noise or bandpass filters.
- Last not least: code an interface that lets you test the classifier on the spot.